Session II Gut microbiome and xenobiotics
In this guide
In this guideOn this page
Skip the menu of subheadings on this page.Session II Gut microbiome and xenobiotics
The interaction of pharmaceuticals with the gut microbiome
72. Professor Kiran Patil (MRC Toxicology Unit) introduced his talk about the interaction between pharmaceuticals (small molecule pharmaceuticals) and the gut microbiome, which focused on the use of xenobiotics and small molecule drugs and how they can affect the human gut microbiome. Furthermore, how we can map and measure these interactions, disruptions of the microbiome and how this affects design of pharmaceuticals.
73. The speaker introduced mapping drug interactions, which are all affected by diet, age, routine and other factors and can make a huge impact on the microbiota. This makes studies very difficult to design. The bottom-up method is used for designing these studies. This is done by simplifying the problem by reducing the number of factors to 2 and determining how these interact and then expanding this is include greater complexity between all the factors.
74. The hope is that the bottom-up approaches will meet the top-down approaches to give a full insight of the microbiome structure. So far this has been achieved by cultivating bacteria in vitro in defined growth media to study their mechanistic action.
75. One study example showed the impact of a non-antibiotic drug on human gut bacteria. The Prestwick Chemical Library, which contains information about thousands of compounds, was used and the results were expressed as a heat map showing the damage of different drugs on microbiome bacteria. There was a huge impact from antibacterial compounds, as expected but also of non-antibacterial human targeted drugs and the disruption that these cause to the microbiome.
76. Interestingly, >24% of nonantibiotic drugs inhibit the growth of at least one commensal species; the drugs were in a broad range including non-steroidal anti-inflammatory drugs and antipsychotics. This is still expected to an underestimate, as not all of the compounds were evaluated so the impact of pharmaceuticals is anticipated to be huge.
77. The effects of human-targeted drugs were more specific versus those of antimicrobials which were more widespread. However, human-targeted drugs affected abundant commensal species so may have a broader effect on the gut microbiota physiology.
78. The speaker then discussed the effects of non-antibiotic drugs on the gut microbiome. In vitro studies showed drug effects on the gastrointestinal tract which are very similar to those seen with antibiotics. There were some limitations in these studies due to effects such as diarrhoea.
79. An association that has been identified, is the sensitivity of a microorganism to both antibiotics and non-antibiotic compounds. This suggests that resistance to antimicrobials can be exacerbated by the presence of non-antimicrobial compounds and bacteria can become very sensitive to these (Maier et al., 2018).
80. A prospective Japanese cohort study suggests antibacterial resistance genes increase in polypharmacy and showed distinct effects of multiple medications on the human gut microbiome (Nagata et al., 2022). Polypharmacy was also associated with changes in microbial functions, including the reduction of short-chain fatty acid metabolism and increased bacterial stress responses. Even non-antibiotic drugs were significantly correlated with increased antimicrobial resistance potential through polypharmacy.
81. Positive and negative interactions that can occur between specific bacteria and different pharmaceuticals that they come into contact with is called the drug-bacteria interaction network.
82. Metabolite secreted by bacteria strongly interact with intracellular proteins and have the capacity to cause cascading effects on the immediate environment of the bacteria. A change in metabolite secretion can dramatically influence the community composition, which can be seen in a synthetic community.
83. The products of xenobiotic biotransformation by bacteria can also affect their secretion of metabolites.
84. Emergent phenotypes involve the cross-protection of different microorganisms when undergoing sensitisation following exposure to drugs. This particular vector in the community through bio-accumulation or biodegradation, or sometimes using both mechanisms, actually protects other members of the community in the microbiome.
85. Attention was drawn to the impact of common chemical pollutants on the gut bacteria, which includes environmental contaminants, pesticides, and food contact materials. An image was presented of the pollution library which contains more than a thousand different compounds, including pesticides and industrial chemicals. This library shows the broad ranges of chemicals that can affect the human microbiota and the wide range of effects they can have on the bacteria and how this is comparable to the effects of human-intended pharmaceuticals.
86. The importance of viewing these chemicals from a biological perspective, and not only from the chemical perspective, is critical when categorising them and considering how they need to be controlled to prevent bacterial resistance, and other damage.
Potential adverse effects of pesticides on the microbiome
87. Mr Neil Lister (Crop Life International Representative) firstly introduced the WHO/FAO Joint Meeting on Pesticide Residues (JMPR) who have begun requesting available data on the impact of pesticide residues on the human gut microbiome when recommending Codex Maximum Residue Limits and proposed testing using a guideline developed for antimicrobial veterinary medicines.
88. Crop Life’s main questions are surrounding: the outstanding safety questions relating to potential effects of pesticide residues in food on the human gut microbiome and how well advanced is the scientific understanding of the human gut microbiome and what is the likelihood of pesticide residues in food, at the concentrations at which they are present, having an adverse effect?
89. The speaker then discussed what information is already available for pesticides. Each pesticide active ingredient is assessed by generating a large mammalian toxicity dataset (acute toxicity, genotoxicity, neurotoxicity, developmental and reproductive toxicity, and repeat dose systemic toxicity (short and long term)), mostly obtained using oral dose in-vivo studies. Adverse health outcomes that may be mediated by the gut microbiome have likely been assessed and incorporated into the health-based guidance values derived from these data, albeit indirectly.
90. A current data gap is specific experimental data on the impact of pesticides directly on human gut microbiomes. These datasets are rarely available (although they may exist as historical data) and are not currently required by regulatory agencies.
91. In summary, further science and development is needed to investigate how pesticides impact the human gut microbiome and its influence on human health, specifically in:
- Defining a healthy microbiome and understanding normal fluctuations in the microbial composition.
- Understanding how changes in the microbiome relate to adverse health effects.
92. Finally, the speaker prosed some questions that will need to be addressed. including what additional mammalian hazards are not addressed by current data? and if specific experiment data for a pesticide are required, a globally harmonised and validated test guideline (ideally via the OECD) should be developed.
Microbiological Risk Assessment of Residues of Veterinary Medicines
93. Dr Silvia A. Piñeiro (U.S. Food and Drug Administration (FDA)) introduced her talk which focused on how new animal drugs are assessed and the importance of their effects on the human intestinal microbiota.
94. In traditional toxicology, an assessment is performed to establish an Acceptable Daily Intake (ADI) and determine a safe concentration for veterinary drug residues in food. The ADI is an estimate of the amount of a substance, expressed on a body weight basis, that can be ingested daily over a lifetime without appreciable risk to human health.
95. For antimicrobial drugs, a human intestinal flora assessment needs to be performed to determine whether a microbiological ADI (mADI) is necessary (Piñeiro et al., 2021).
96. The concern for antimicrobial residue effects on the human intestinal microbiome is that the human intestinal microbiome is a complex group of microorganisms composed of bacteria, fungi, archaea, protozoa, and viruses, organized in a community, living in close relationship with their host and the human gastrointestinal tract environment. This is a balanced, diverse, and complex ecosystem performing essential functions that may be disrupted by drugs and their residues.
97. In vivo and in vitro systems have shown that very low levels of drug residues from edible animal tissues can alter the intestinal microbiome and may result in human health consequences.
98. Drug residues present in edible animal tissues may reach the human colon by the oral route due to incomplete absorption and may be absorbed, circulated, and excreted via bile and/or secreted through the intestinal mucosa.
99. United States developed guidance for industry (GFI) #159/VICH GL36 and first implemented the guideline in 2005. It describes the approach for establishing mADI’s.
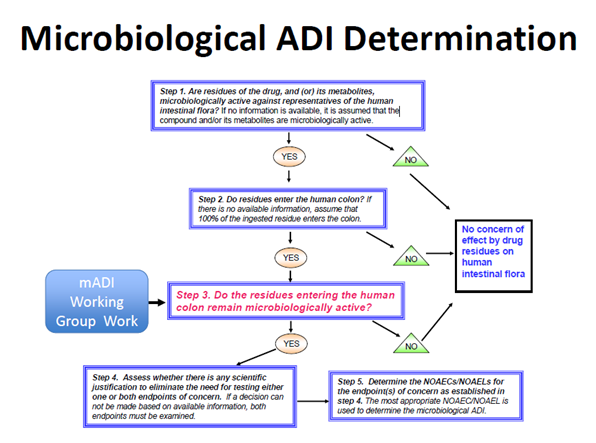
100. A five-step decision tree illustrates the approach for determining whether drug residues reaching the human colon remain microbiologically active, and whether a mADI determination would be necessary for drug approval (Figure 1).
Figure 1. Microbiological ADI Determination. Figure taken from Piñeiro, S.A. and Cerniglia, C.E., 2021. Antimicrobial drug residues in animal‐derived foods: Potential impact on the human intestinal microbiome. Journal of Veterinary Pharmacology and Therapeutics, 44(2), pp.215-222.
101. It recommends in vivo or in vitro test systems and methods for determining no-observed adverse effect concentrations/levels (NOAEC/Ls) for endpoints of human health concern and a procedure to determine a mADI from the NOAEC/Ls. The endpoints of concern are: disruption of the colonization barrier and development of antimicrobial resistance.
102. When establishing the mADI, the microbiological endpoint with the lowest value is used; either disruption of the colonisation barrier or an increase in the population(s) of resistant bacteria.
103. There are multiple approaches to addressing the impact of residues on the human intestinal flora based on Veterinary International Conference on Harmonization (VICH) GL36.
104. Six mADI examples were presented based on a variety of microbiological endpoints: tilmicosin (disruption of colonization barrier); narasin (disruption of colonization barrier); lincomycin (disruption of colonization barrier); tetracycline (Increase in the population of resistant bacteria in the human colon); amoxicillin – Joint FAO/WHO Expert Committee on Food Additives (JECFA, 2017).
105. For amoxicillin, a microbiological ADI of 0-0.002 mg/kg bw could be established on the basis of disruption of the colonization barrier of the gastrointestinal tract and using the adopted colon content volume of 500 mL. For ampicillin reviewed by JECFA in 2017, the overall mADI of 0-0.003 mg/kg bw was based on the increase in the population of ampicillin-resistant bacteria in humans, and using a safety factor of 10, as this microbiological end-point is lower than the microbiological ADI for its effects on colonization barrier disruption.
106. The GFI #159/VICH GL36 was later revised after new research and discussions.
107. The addition of Appendix D in 2013 ‘Regarding the Determination of the Fraction of Oral Dose available to Microorganisms’ was a key step forward in improving the guidelines. The change of the colon content volume to 500 mL/day, implemented in the US in 2022 was also highlighted.
108. Finally, the speaker pointed out the data gaps which include:
- what can be considered a normal microbiome?
- what are the most sensitive indicators/biomarkers of microbiome dysbiosis and how do single and combined toxic exposures effect the intestinal microbiome?
- Metagenomics vs metatranscriptomics vs metabolomics?
- What about the applications of new methodologies for establishing a mADI?
- What are sensitive indicators/biomarkers of dysbiosis?
- Changes in populations? Changes in microorganism functions (metabolomics, immunity?)