Gut Reactions: Xenobiotics and the Microbiome Workshop Report London, UK 2024
Gut Reactions Workshop
In this guide
In this guideBackground and Objectives
In this guide
In this guideBackground and Objectives
1. A wide range of substances in the diet come into contact with the intestinal microbial community (the gut microbiome) and therefore have the potential to influence this community and sometimes, in turn, host health. The implications that this has for the assessment of the risks to human health of chemicals and other components of the diet and how this can be determined are currently under debate; as yet there is no clear consensus.
2. In this workshop we set out to explore the complex current state of the science of the microbiome pathophysiology and the possible impact of xenobiotics on host-microbiome interactions and vice versa, including possible mechanisms and health implications, with a particular emphasis on the gut microbiome and dietary exposure.
3. The workshop output will hopefully enable new insights, provide a review of the science, initiate discussions to determine where the data gaps are in research, what effects are of concern, and how might xenobiotics be evaluated practically for such effects in the future.
Workshop Overview
In this guide
In this guide4. The workshop took place in October 2024 in London, United Kingdom. The workshop included an overview, themed sessions consisting of short flash presentations followed by roundtable discussions. There was worldwide attendance from multiple stakeholders including academia, government and industry.
5. The four sessions were:
Session I: Interactions of the host microbiome system
Session II: Gut microbiome and xenobiotics
Session III: Assessing the impact on the microbiome
Session IV: Future Directions
Introductions and aims of the day
In this guide
In this guideIntroductions and aims of the day
Overview
Introduction to the microbiome: The gut microbiome and food safety
6. Professor Gant (UK Health Security Agency) introduced his talk by acknowledging the previous work of the COT in this area, which is available on the COT website (Statement on interactions between xenobiotics and the human microbiota and their potential toxicological implications.pdf).
7. The speaker then provided an overview of the gut microbiome in relation to toxicology and food safety.
8. The community of bacterial, viral and fungi microorganisms (called symbionts) is classed into three categories: mutualists (benefit themselves and the host), commensals (benefit themselves but not the host (but do not harm the host)), and pathogens (benefitting themselves but harming the host).
9. In addition to the alimentary canal, such communities are found on any body surface that has a connection with the environment and particularly where conditions are favourable to microbial growth: skin, oral cavity, vagina, lungs.
10. The total diversity of the microbiome is probably about 100 trillion organisms of which we have identified about 1% or less. The gene pool far exceeds that of the host.
11. There are approximately 1012 bacteria in 1g of human faeces. The human gut microbiome is thought to be about 8% viral sequences with about 1011 viruses per 1g of stool.
12. The number of bacteria in the body is actually of the same order as the number of human cells, and their total mass is about 0.2 kg (Sender et al., 2016). Previously, a ratio of 1:10 for human cells to bacterial cells has been widely reported. New calculations suggest equal bacterial cells to human cells in all of us (approximately 3x1013 cells).
13. Therefore, it has been suggested that the number of all organisms in the gut exceeds that of human cells in the whole body (Sender et al., 2016), and that this number can be affected by the host’s diet and other factors such as age, lifestyle and intake of therapeutics. The type of symbionts present can also change, leading to an unbalanced gut microbiota or dysbiosis.
14. It was noted that the gut microbiome interacts with other organs such as the brain and liver i.e. forming a gut/brain axis and gut/liver axis. Examples of metabolic reactions by the gut microbiome were briefly introduced, some of which are specific to the gut microbiome. These specific reactions can have great importance to risk assessments and can often influence adverse outcome pathways (AOP).
15. The speaker concluded by posing some questions:
- How does our environment affect our microbiome?
- How do genetics affect the microbiome?
- How does the microbiome affect phenotype?
Microbiome manipulation- Government Office (GO) for Science UK Government Review
16. Dr Chrysi Sergaki (Medicines and Healthcare products Regulatory Agency) presented on the outcome of the GO Science UK Government Review roundtable discussion held in April 2024 on Microbiome Manipulation via diet, pre-/pro-biotic and other interventions including research gaps.
17. An overview was given highlighting that we are only half human, having more microbial cells than human cells in the body, and cells in the gut are associated with many functions of the human body. Where the balance in the gut microbiome is disturbed, it has been associated with many serious diseases and conditions, such as Parkinson’s disease, as well as musculoskeletal, digestive and pulmonary conditions.
18. The presentation then moved on to interventions, which included the discussion at the roundtable that acknowledged the microbiome is highly complex and varied between individuals. It was discussed that we are not able to define “healthy” and “unhealthy” microbiomes as these can look different across different people and gut microbiome compositions. However, it is known that a "healthy" microbiome should be well balanced (both in terms of diversity and co-existence of species) and is associated with the presence of specific bacteria.
19. Pre-biotics and pro-biotics may have different, and sometimes minimal, effects on the gut microbiome due to variability in the microbiome, differences in diet and other characteristics among individuals. There are also variations in biological responses to probiotics among individuals. Actions should be taken to increase public/consumer awareness of these considerations so they can make more informed decisions.
20. It was highlighted as a concern that when the role of the microbiome is discussed in the mainstream media it may not be supported by evidence and public awareness needs to be raised.
21. It was stated that bacteria that are considered to be beneficial can also have negative impacts on health. However, these rare negative impacts should be weighed against the many positive cases of improved health and reduced disease.
22. It was suggested we need to move towards studying the microbiome and its function / interaction in the body rather than focussing on just taxonomy.
23. The presentation moved on to products with pre- and pro-biotic effects that are currently on the market, however, these can have different effects on different people with varying biological responses. Fermented foods are newly available products, but the literature is sparse and there is concern over antimicrobial resistance (AMR) burden. Not one size fits all, so these products can have different effects depending on an individual’s characteristics and differences in diet. The effects can vary with the number of microbial organisms in probiotics and once intervention is stopped the microbiome can go back to its original state.
24. When administered responsibly, faecal microbiota transplantation (FMT) has been shown to help reinstate diversity in the gut microbiome. Use of FMT is regulated in the UK but more research is needed to increase its range of applications.
25. It was noted that knowledge on the microbiome and its importance to human health has been available for over 15 years so why do we not have more products? One of the main issues faced is the lack of consistency in testing/studies which limits the validity of claims, so standardisation is needed. Studies focus mostly on the United States (US) population or other western populations so there is a need for other populations to be included.
26. The research gaps were discussed, and these included the need to define the baseline microbiome e.g. what does the microbiome look like before any interventions to the core diet. Clinical studies have been inconsistently recorded, metabolites and other biomarkers can be used but it is difficult to take accurate measurements, methods for retrieving samples also need improving as currently they are invasive or based on stool samples, while animal studies do not translate well to humans, especially when looking at mechanisms of action.
27. The presentation concluded with current regulatory considerations and what can be done to help to facilitate innovation in the field and to enable translation to products. Suggestions included the need for clear clinical research guidance, especially for dietary pro-biotic and pre-biotic interventions. There are currently a few grey areas when it comes to some products e.g. the borderline between medicine and food and sometimes these products can potentially be covered by two different agencies (MHRA for medicine and Food Standards Agency (FSA) for food), so clarity is needed.
Session I Interactions of the host-microbiome system
In this guide
In this guideSession I Interactions of the host-microbiome system
Food additive emulsifiers and their impact on gut microbiome, permeability, and inflammation: Mechanistic insights into inflammatory bowel disease metabolomic and gut microbiome profiles & Maternal and early child health, women's health and the gut microbiome as a modifiable factor with nutrition to improve health outcomes
28. Dr Federica Amati (Imperial College London and ZOE) introduced the microbiome as an ecosystem / garden which helps keep the body healthy but can also contribute to illness as well as impact health outcomes. The speaker then presented on why the microbiome is important including effects on mood; appetite; food choices; influences on the immune system; impacts on the menopause and bone health; it breaks down fibre and polyphenols and produces postbiotics (enzymes, vitamins, fatty acids) (Valdes et al., 2018). Some preliminary studies have shown that modulating the microbiome can improve health outcomes.
29. The speaker then stated that the relationship between our microbes and food are complicated, complex subjects. In inflammatory bowel disease (IBD, which includes Crohn’s and ulcerative colitis) gut permeability increases. allowing bacteria and toxins to cross the gut barrier which triggers an immune response. This immune response promotes chronic inflammation and results in worsening symptoms.
30. Gut dysbiosis is an imbalance of beneficial vs harmful bacteria and plays a significant role in IBD by weakening the gut barrier. This increases the permeability of the gut barrier and exacerbates inflammation. A reduction in the number of beneficial bacteria like lactobacillus and bifidobacterium is common.
31. Research has shown that IBD patients have distinct gut microbiome profiles with lower diversity and fewer beneficial bacteria. Pathobionts (harmful bacteria that thrive in dysbiosis), such as Enterobacteriaceae and Fusobacterium, are often elevated in IBD, driving inflammation. Tight junction dysregulation is a situation where the structures maintaining gut barrier integrity become compromised in IBD leading to a “leaky gut”. Proteins like occludin and zonulin regulate these junctions. In IBD, pro-inflammatory cytokines are overproduced due to a dysfunctional immune response to gut bacteria. These cytokines disrupt the gut barrier perpetuating inflammation and gut permeability.
32. Gut bacteria produce various metabolites such as short chain fatty acids (SCFAs) and bile acids. SCFAs strengthen the gut barrier and regulate inflammation. Disruption of these metabolites in IBD and other gut disorders leads to impaired gut function. Metabolic analysis of IBD patients shows reduced production of SCFAs especially butyrate, which normally helps maintain gut integrity. Butyrate deficiency contributes to increased gut permeability and inflammation.
33. Studies indicate that the western diet (high in protein, ultra processed foods (UPF) and sugar) has led to a 50% decrease in the diversity of the microbiome. This was determined by a comparison with the African Hadza tribe whose microbiome is more diverse than the western population. It was noted that further investigation of the microbial diversity of the microbiomes of other cultures is needed.
34. When considering the impact of emulsifiers and food additives on the gut microbiome the entire dietary pattern needs to be considered. In the UK, greater than 50% of the average person’s diet is made up of UPFs. People are consuming significant amounts of novel foods containing combinations of chemicals which humans have not evolved to consume. Food additives and emulsifiers should be considered as a mixture and not stand-alone components. As a result, there is a lot of information, and unknowns to take into account when assessing the risk they pose.
35. Studies have shown when comparing high to low UPF diets there is a trend towards reduction in gut microbiome diversity and negative metabolic health effects in high UPF diets. Less healthy food patterns gave rise to a less healthy gut microbiome, and this was associated with adverse metabolic effects. When considering artificial sweeteners, the science is varied and whilst it is widely accepted that they don’t enter the blood stream they do have local effects on the microbiome; in addition, food colourants have been linked to gut dysbiosis.
36. There is evidence to suggest that emulsifiers can lead to inflammation and that diets lower in emulsifiers are linked with fewer irritable bowel symptoms in humans. Emulsifiers have been shown to disrupt the lumen of the gut wall leading to leaky gut. This allows the movement of the components within the gut lumen to “leak” out.
37. Furthermore, there is some evidence that people who wash dishes and don’t rinse off the soap (which is an emulsifier) have a higher incidence of colon cancer due to gut microbiome disruption.
The maternal and infant microbiome
38. The second topic of discussion presented was that of maternal and early child health, women's health and the gut microbiome as a modifiable factor, using nutrition to improve health outcomes (Leeming et al., 2019).
39. During pregnancy, the gut microbiota shifts to support fetal growth. Dysbiosis in pregnant women is linked to adverse outcomes like gestational diabetes, preeclampsia, and preterm birth. A healthy maternal microbiome supports optimal pregnancy outcomes.
40. The maternal microbiome strongly influences the infant’s gut microbiome, especially during vaginal delivery and breastfeeding.
41. It is recognised that the gut microbiome is very malleable in the first 3 years of life, and early gut colonisation shapes the child’s immune system, digestion, and risk of developing allergies or chronic diseases later in life.
42. Research is being undertaken with a view to improve the microbiome of babies who are formula fed.
43. Gut and vaginal microbiome dysbiosis can have a negative effect on fertility and pregnancy outcomes. Tests are now available to diagnose vaginal dysbiosis and to predict the likelihood of preterm birth.
44. Observational studies have shown a relationship between babies growing up in spaces with pets and green spaces to have an improved microbiome and fewer allergies in later life.
45. Increasingly children in the UK are being diagnosed with poor health outcomes, including early adiposity, poor mental health and increased respiratory issues, which was correlated with UPFs (Oliviera et al., 2022).
46. The gut microbiota affects key aspects of women’s health including hormonal balance (e.g. oestrogen metabolism) and conditions including polycystic ovary syndrome, endometriosis and menopause. Dysbiosis can worsen these conditions by influencing inflammation and hormone regulation.
47. It has been suggested that the microbiome plays a role in the menopause transition and health. Studies on pre, peri and post-menopausal women have shown marked differences in responses and evidence shows that the markers measured were mediated by the gut microbiome composition. The marker levels measured in these women were more similar to those seen in men. A higher quality diet (which was considered to be one high in whole foods and mostly plant based) led to a lower prevalence of menopausal symptoms. Diet quality is associated with menopause symptoms. There was also a marked difference in the aspects of the gut microbiome that were measured due to this diet.
48. The Gut-Immune axis was introduced, with the speaker stating that the trillions of microbes in the gut train and educate the immune system, building defences, keeping the peace and ensuring optimal function. Therefore, looking after gut health is one of the easiest ways to look after immune health. This was followed up with evidence that increasing fibre intake by 5 grams per day could reduce unwanted inflammation (Shivakoti et al., 2022).
49. Finally, the speaker also discussed a concept called “proprietary gut microbiome scoring” which uses metagenomic gunshot sequencing integrated with MetaPhlAn4 (a computational tool for profiling the composition of microbial communities) to allow the comparison of microbiomes. The aim is to link certain microbial strains with specific health outcomes. Preliminary data shows that there is a significant difference in the microbiomes of US and UK cohorts when using this scoring method.
Antimicrobials in livestock and the potential impact on gut microbiota and colonisation by zoonotic pathogens
50. Professor Rob Kingsley (Quadram Institute) firstly described how antimicrobials are given to animals in agriculture to treat infections or as growth promoters.
51. The talk then focused on how the gut microbiota prevent pathogen invasion and how disruptions to the microbiota lead to pathogenic bacteria growing to high levels in the intestines of livestock.
52 .The speaker then explained colonisation resistance during homeostasis (Rogers et al., 2021). Bacteroida and Clostridia are important for the colonisation resistance of the gut from pathogens. They produce SCFAs such as propionate, acetate and butyrate, which can inhibit Enterobacteriales. Butyrate drives oxidative phosphorylation in epithelial cells, which are the main entry route for xenobiotics in the gut. Oxidative phosphorylation consumes oxygen and drives epithelial cell hypoxia and creates an anaerobic environment (anaerobiosis). The anaerobiosis maintains homeostasis and the abundance of obligate anaerobes, such as Clostridia and Bacteroida, and low levels of Enterobacteriales.
53. The speaker then discussed the impact of antibiotics on gut bacteria. Antibiotic use leads to depletion of Bacteroida and Clostridia in turn leading to depletion of SCFA and butyrate levels. Depletion of butyrate results in a switch to aerobic glycolysis which in turn causes oxygenation of the mucosal surface; inhibition of obligate anaerobes and expansion of Enterobacteriales (Rivera-Chávez et al., 2017). This is further compounded by the increase in carbon sources such as sialic acid, fructose, galactarate, which can be used by Enterobacteriales. AMR is another problem in this situation as Enterobacteriales can become resistant to the antibiotics and further outcompete any commensals.
54. The speaker then discussed some of the work on a high copper diet in post-weaning piglets. Piglets are fed these diets as copper is used as a growth promoter and it reduces post-weaning diarrhoea. There is concern that pathogens are becoming adapted to the copper and there is co-selection for AMR. There are further concerns that co-resistance in pathogens as a result of copper use can lead to microbiome modifications as it could allow pathogens to become adapted to a different niche, normally occupied by commensals.
55. Epidemiology work identified the emergence of Salmonella typhimurium ST34 copper resistant strain (Branchu et al., 2018). This is one of the main foodborne pathogens. This resistant strain is thought to have emerged due to adaptations to the changing environment of the host. Resistance genes in this bacterium increase the minimal inhibitory concentration (MIC) of copper, especially in anaerobic conditions.
56. A farm study conducted in piglets that were fed high and low copper diets showed small changes to the microbiome with increased abundance of 4 species and decreased abundance of 10 species in response to the high copper diet. In the metabolome, 11 out of 67 metabolites were altered by copper. Butyrate, propionate and acetate levels were unchanged in the high copper diet, but formate had increased.
57. Therefore, the effect of a high-copper diet on colonisation resistance showed no significant reduction in Clostridia and Bacteroida and no reduction in short chain fatty acid production. The increase in formate, was a cause for concern with respect to the potential expansion of Salmonella. In addition, the decreased relative abundance of commensal Enterobacteriales might decrease the competition for Salmonella.
Effects of oral iron supplementation on the gut microbiota / different responses between the sexes to increased dietary protein on the gut microbiota using pig models
58. Dr Marie Lewis (University of Reading) presented a talk on the effects of oral iron supplementation on the gut microbiota, and the different responses between the sexes to increased dietary protein on the gut microbiota, using pig models.
59. The speaker discussed two research projects:
- Do high-protein diets have the potential to reduce gut barrier function in a sex-dependent manner?
- The effects of iron supplementation during infancy.
60. The first project looked at the sex-dependent effects of high protein diets on gut barrier function (James et al., 2024). The speaker noted that there is currently considerable promotion of high dietary intakes of protein and increased availability of high protein bars and foods. Also, higher protein intakes are encouraged in older age but are not effective without fibre consumption and exercise.
61. The speaker noted that it was previously thought that protein was completely absorbed from the gut but, around 10% reaches the colon, where it can be utilised by gut bacteria. The gut bacteria utilise oligosaccharides and fibre, and when the gut bacteria utilise protein, there is a shift in their metabolic profile. This metabolic shift increases the amount of negative microbial end products, such as ammonia, p-cresole and indole. These end products have been associated with reduced tight cell junction proteins which can potentially cause a leaky gut. The higher amounts of microbial derived end products enter the bloodstream and can lead to inflammatory responses. Therefore, the speaker indicated, there is a theoretical link between high protein diets and long term chronic inflammatory conditions such as coronary heart disease.
62. It was highlighted that the gut microbiota and the immune system are different between males and females. Therefore, it is logical to assume that the effect of different substances on the gut microbiota will vary between sexes.
63. The research project was initiated with a screening process using in vitro gut models. Types of animal protein analysed included: whey, milk, fish and eggs. It was noted that a lot of other research in this area uses very concentrated hydrolysed proteins. However, the issue with using hydrolysed proteins is that they are almost completely absorbed in the small intestine. Therefore, use of gut model systems would be artificial as the protein would not reach the colon. To prevent this occurrence in the study, the research group extracted only 70% of proteins so the substrates they were testing contained other substances that individuals would consume if they had a high protein diet.
64. The main observation from this study was a change in the gut microbiota, with some populations of bacteria being affected whereas others were not. Additional observations were that the type of protein influenced the type of microbial end products produced. Therefore, the speaker suggested, advice on increasing protein in the diet should specify the type of protein. The speaker found that analysing the output of the microbiota (i.e. the end products) is more important than the composition of the microbiota.
65. Further results from the study were that there were sex differences in response to different types of proteins. Therefore, future advice on dietary protein intakes may have to vary between males and females. The latter part of this research on the effects of high protein diets on gut barrier function utilised a pig model. The speaker explained that the pig model was selected due to issues with translating results from rodent models to humans. Rodents live in a high hygiene facility, which alters their microbiome and leads to an unusual development of their immune system and metabolism. However, pigs are more similar to humans in terms of their gut physiology and microbiome and their inter-interindividual microbiome variation.
66. Both male and female piglets received either a low protein diet or a high protein diet (i.e. 28% protein) totalling four treatment groups. Results were that groups fed the higher protein diet grew faster, although there was no difference in growth rates between males and females between weaning and 12 weeks of age. Other findings were that the same differences in metabolic profile observed in the former part of the study were also observed in the latter study with piglets. The speaker concluded that the study provided direct evidence that high protein diets reduce the expression of gut barrier-function proteins in a sex-dependent manner (James et al., 2024).
67. The speaker then discussed the project on iron supplementation during infancy, which was conducted as infants fed on formula milk are receiving high amounts of iron. The speaker acknowledged that iron deficiency in infants is a potential concern but noted that, similarly to protein, high amounts of iron in the colon occurring due to lack of absorption from the gut can cause a shift in the metabolic profile of microbiota.
68. The speaker explained that piglets were received at one day of age and were not bottle fed but received formula milk from trays. It was noted that if piglets do not consume iron, they will develop anaemia in 2-3 weeks however, rodent models require a longer period before they develop anaemia.
69. The initial findings were that all types of iron supplementation were able to prevent anaemia. However, the control group were the only piglets that grew normally, and the groups receiving oral iron had a reduced growth rate similar to that of anaemic piglets. These findings implied that administering formula milk containing iron could be reducing the growth rate of infants. Additional findings were that there were no differences in host metabolites between any of the iron treated groups. Furthermore, administration of oral iron had an impact on the composition of the microbiota. The speaker highlighted the associations found between weight gain and different members of the gut microbiota. The researchers were able to identify bacteria associated with increased weight and bacteria associated with decreased weight.
70. In the oral iron group, bacteria present in the microbiome had negative associations with weight gain (i.e. decreased weight gain). Final results indicated a direct link between specific components of the gut microbiota and lack of weight gain in infant piglets as a result of iron supplementation.
71. The main conclusions were that for the first time, in an animal model that is reflective of humans, protein was shown to directly impact gut barrier function in a sex-dependent manner. Also, females may be more prone to reduced gut barrier function following high protein diets compared to males. Furthermore, different types of protein have different effects on the composition of the gut microbiota and metabolic profiles of microbial-derived end-products. Finally, iron supplementation resulting in reduced weight-gain was associated with specific components of the microbiota that are less abundant.
Session I Roundtable Summary
In this guide
In this guideSession I Roundtable Summary
Can a healthy gut microbiome be defined through quantifiable characteristics? and how does that change? Through life changes? i.e. Maternal, sensitive populations and variability
- It was acknowledged that there was a lack of a concrete definition of the microbiome and a specification, or an ‘average’ characterisation, of a healthy gut microbiome is not yet able to be established. Global harmonisation of a potential definition would prove challenging, as varying factors will need to be considered including different life stages, disease states, ethnicity, diet etc. Due to the complexity of the topic, there was acknowledgement that this may not be as simple as a definition but rather a specification or guidelines containing parameters, ranges, diversity and species information. It was noted that what may be considered a ‘healthy’ gut microbiome for one individual may not be the same for another.
- Although it was noted that every microbiome is unique to each individual; by using a comparison with a large database of microbiomes it would allow researchers to see if there were any common factors for a ‘good’ microbiome.
- Identify a baseline: what does the microbiome look like before any interventions to the core diet.
- Defining the principles that we apply in a risk assessment setting could take into account age, sex, genetics, disease effects/changes.
- We should move away from trying to establish a definitive healthy/normal microbiome due to inter-individual variability, and factors affecting this (diet, lifestyle, health status). We should try to define ‘reference populations’ with less dependence on an individual’s health status, rather incorporating the contextual factors mentioned above, such as diet.
- It needs to be determined whether a change in the microbiome is a marker of a separate causative event or if the change in the microbiome is the cause itself. It is possible to provide evidence in both directions, which increases complexity. In this way, the microbiome will adapt in terms of population composition as a result of a sustained dietary change, whereas if the change is temporary, only the metabolic activity of the current population will change. Therefore, linking a microbiome change to a marker is complex. Additionally, when considering “unhealthy” markers, are they reactive to an effect or pre-emptive of one? It was suggested that research conducted on a single family could help identifying these markers.
- Bacteria are not the only constituent of the human microbiome, e.g. there are also fungi and yeast, and thus they should also be considered in research.
- With the increase in genomic sequencing of bacteria in the microbiome, it was suggested that it is important to establish the phenotypic properties as well as the genotypic profile of the microbiome, particularly as these are not always correlated and could be impacted by external factors other than just the genetic makeup.
- It was commented that different species can provide different functions, but it is not always clear which species are responsible. Once these functions are further understood there may be a better understanding of the microbiome as a whole. Some species can be ‘good’ or ‘bad’ dependent upon circumstances and surroundings.
- State what model organisms are available to researchers that accurately reflect the behaviour of bacteria found in the gut microbiome.
Main Themes
- “Healthy” microbiome not yet defined i.e. a baseline.
- Specification, or an ‘average’ characterisation.
- Guidelines containing parameters, ranges, diversity and species information.
- Should try to define ‘reference populations’.
How much variability in an individual’s gut microbiome is normal, and how resilient is the microbiome to change?
- It was noted that as there is so much variability it would not be possible to tell by looking at the microbiome itself what is good and what is bad, instead the focus should be on looking at the adverse health outcomes.
- Underlying socio-economic factors are some of the biggest determinants of individual health outcomes.
- Microbiome changes dramatically over a lifetime.
- UK Biobank may provide the variability of an individual’s gut microbiome.
- Attendees stated that an adaptation of the microbiome is not necessarily beneficial, it depends on the host-microbiome interactions. For example, a certain microbiome population in a specific individual will have no immune system consequences, whereas the same population in a different individual will result in adverse effects.
- It was discussed that there is a need for better quality microbiome data, suggesting that, as there is not yet a defined link between the microbiome and adverse outcomes, that adverse outcome pathways (AOPs) are required to strengthen the evidence base.
- Rather than studying the microbiome in isolation, microbiome analysis could be incorporated into other studies as supporting information. Emulsifiers have been identified as a possible cause of adverse effects, and it was proposed that these could serve as a case-study for risk assessment.
- It was noted that it is the long-term impact on the microbiome that is important. While antibiotics have a large impact, they are mostly used short-term. However, other medicines that are used long-term also have effects on the microbiome, e.g. statins and proton pump inhibitors. It was stated that hundreds of pharmaceuticals administered for other purposes have antimicrobial effects.
- It was questioned whether having a less diverse microbiome has a negative impact on health outcomes. It was also questioned whether individuals could decide for themselves how healthy they felt.
- Defined principles may have to be applied to given populations and will vary according to age and sex.
- The resilience and/or stability of an individual’s microbiome may also be an indicator of health state. It was further suggested that laboratories performing studies in humans could compile a database to help define ‘normal’ in a way similar to the collection of historical control data in animal studies.
- Within this, susceptible populations also need to be considered for example, early life. It was further discussed that the microbiome is more malleable at an early phase in life becoming more stable over time. As a result of this, newborns would be a difficult population to monitor.
- It was commented that when considering risk assessment, any change in the microbiome could be suspect and therefore a potential risk, however the challenge is defining what constitutes a meaningful change.
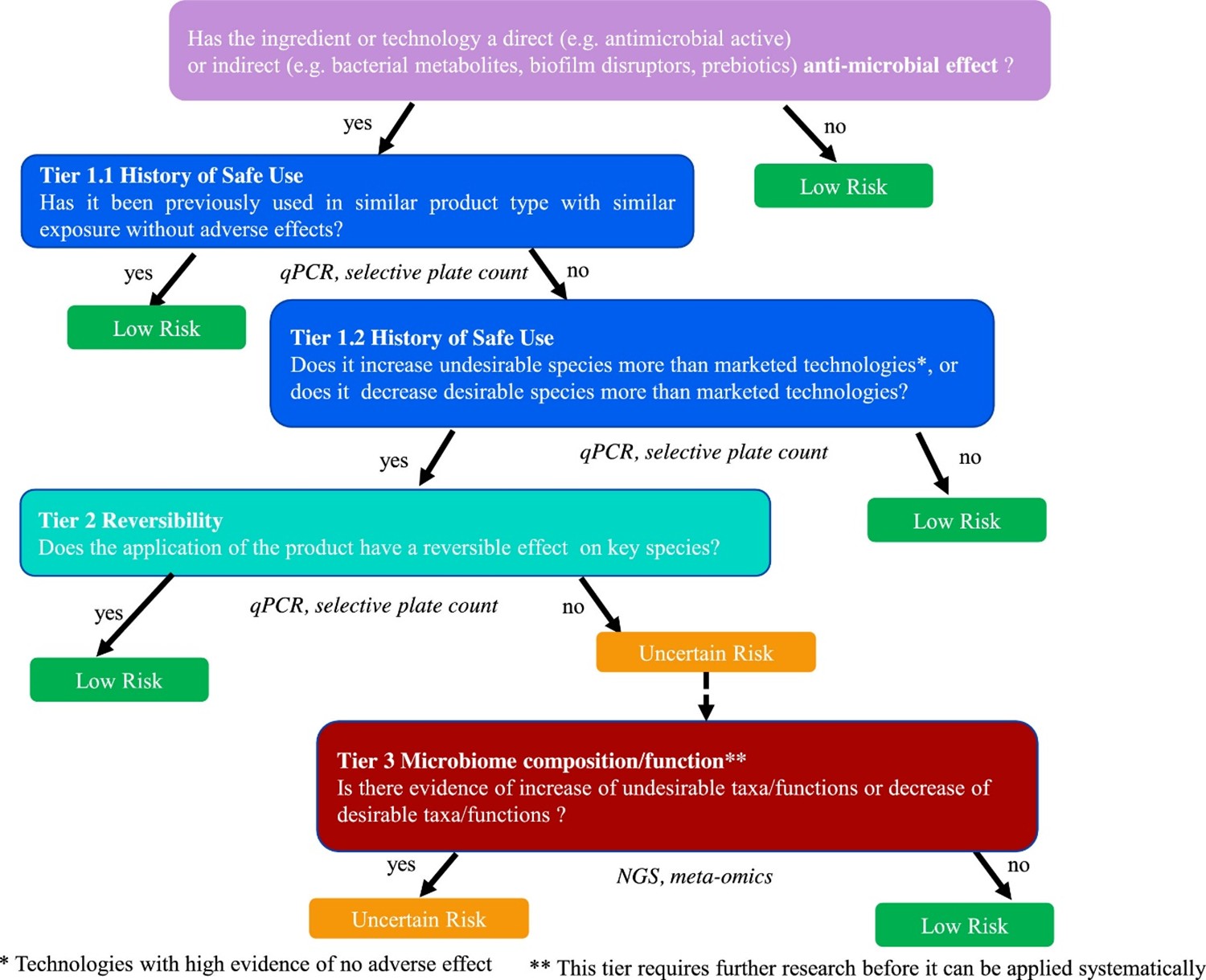
- From a risk assessment standpoint each group/ life stage should be evaluated differently. From current knowledge a cross-control design would not be possible, however it may be possible for change to be predicted in some groups.
Main themes
- Microbiome changes dramatically over a lifetime.
- Microbiome analysis could be incorporated into other studies as supporting information.
- Focus should be on looking at the adverse health outcomes.
- Any change in the microbiome could be suspect and therefore a potential risk, however the challenge is defining what constitutes a meaningful change.
- In the future, may be possible for change to be predicted in some groups.
What is normal, and the complexity of factors contributing to susceptibility i.e. to what extent will the COT need to take account of impact on the microbiome in its toxicity assessments?
- Defining a ‘range’ of microbiomes may be one way forward, but it is unclear whether this should be based on structure, function, or both.
- For a risk assessment: what increases or decreases the risk, and what impact does that risk have.
- One challenge is how to account for the different factors internally and externally.
- Chemical conversions occur in the gut, which have to be taken into account in a toxicological assessment.
- Assume 5-10% of individuals have a deleterious biochemical conversion. Do we acknowledge the 10% and as a precaution cannot approve the chemical or mention the possibility of a side effect as would be the case for the pharmaceutical industry?
- Clinical data is as important as dietary information especially treatment data e.g. antibiotics used, medicines used.
- It was agreed that one of the aims of current scientific research is to establish markers that represent a healthy microbiome. However, linking a microbiome change to a marker is complex, including accounting for any epigenetic changes. Additionally, when considering “unhealthy” markers, are they reactive to an effect or pre-emptive of one? It was suggested that research conducted on a single family could help in identifying these markers.
- It was suggested that research on what types of microbiomes increase the risk of adverse effects was necessary. Additional research may include looking at different populations with different gut microbiomes and observing what populations have more resilience to xenobiotics.
- Consider a toxicological endpoint and a microbiological endpoint and which biomarkers should be used when investigating the microbiota including the possibility of how disruption (e.g. antibiotics) could potentially affect the value of the selected microbiological and toxicological endpoint.
- Susceptible populations must also be considered along with groups such as IBD, IBS patients. Other populations such as infants and children have immature microbiomes. Special considerations and studies need to take place to protect the establishment of these microbiomes.
- Could diversity be considered a barrier in determining these endpoints. There is already so much diversity in ethnicity, age, sex, diet, lifestyle factors anyway, is it even possible to account for all of it. Is there a way that all of these can be a contributing factor and go from there.
- Incorporating and factoring in the risks but also the advantages of understanding how we can change the composition of the microbiome when it comes to regulatory products and scientific advice.
- As it currently stands, it was agreed that the data and knowledge are not yet sufficient to allow for conclusions, however where there is knowledge of function this could be used as a starting point and proceed from there.
Main themes
- Defining a ‘range’ of microbiomes e.g. structure, function.
- Chemical conversions occur in the gut, which have to be taken into account in a toxicological assessment.
- What types of microbiomes increase the risk of adverse effects.
Session II Gut microbiome and xenobiotics
In this guide
In this guideSession II Gut microbiome and xenobiotics
The interaction of pharmaceuticals with the gut microbiome
72. Professor Kiran Patil (MRC Toxicology Unit) introduced his talk about the interaction between pharmaceuticals (small molecule pharmaceuticals) and the gut microbiome, which focused on the use of xenobiotics and small molecule drugs and how they can affect the human gut microbiome. Furthermore, how we can map and measure these interactions, disruptions of the microbiome and how this affects design of pharmaceuticals.
73. The speaker introduced mapping drug interactions, which are all affected by diet, age, routine and other factors and can make a huge impact on the microbiota. This makes studies very difficult to design. The bottom-up method is used for designing these studies. This is done by simplifying the problem by reducing the number of factors to 2 and determining how these interact and then expanding this is include greater complexity between all the factors.
74. The hope is that the bottom-up approaches will meet the top-down approaches to give a full insight of the microbiome structure. So far this has been achieved by cultivating bacteria in vitro in defined growth media to study their mechanistic action.
75. One study example showed the impact of a non-antibiotic drug on human gut bacteria. The Prestwick Chemical Library, which contains information about thousands of compounds, was used and the results were expressed as a heat map showing the damage of different drugs on microbiome bacteria. There was a huge impact from antibacterial compounds, as expected but also of non-antibacterial human targeted drugs and the disruption that these cause to the microbiome.
76. Interestingly, >24% of nonantibiotic drugs inhibit the growth of at least one commensal species; the drugs were in a broad range including non-steroidal anti-inflammatory drugs and antipsychotics. This is still expected to an underestimate, as not all of the compounds were evaluated so the impact of pharmaceuticals is anticipated to be huge.
77. The effects of human-targeted drugs were more specific versus those of antimicrobials which were more widespread. However, human-targeted drugs affected abundant commensal species so may have a broader effect on the gut microbiota physiology.
78. The speaker then discussed the effects of non-antibiotic drugs on the gut microbiome. In vitro studies showed drug effects on the gastrointestinal tract which are very similar to those seen with antibiotics. There were some limitations in these studies due to effects such as diarrhoea.
79. An association that has been identified, is the sensitivity of a microorganism to both antibiotics and non-antibiotic compounds. This suggests that resistance to antimicrobials can be exacerbated by the presence of non-antimicrobial compounds and bacteria can become very sensitive to these (Maier et al., 2018).
80. A prospective Japanese cohort study suggests antibacterial resistance genes increase in polypharmacy and showed distinct effects of multiple medications on the human gut microbiome (Nagata et al., 2022). Polypharmacy was also associated with changes in microbial functions, including the reduction of short-chain fatty acid metabolism and increased bacterial stress responses. Even non-antibiotic drugs were significantly correlated with increased antimicrobial resistance potential through polypharmacy.
81. Positive and negative interactions that can occur between specific bacteria and different pharmaceuticals that they come into contact with is called the drug-bacteria interaction network.
82. Metabolite secreted by bacteria strongly interact with intracellular proteins and have the capacity to cause cascading effects on the immediate environment of the bacteria. A change in metabolite secretion can dramatically influence the community composition, which can be seen in a synthetic community.
83. The products of xenobiotic biotransformation by bacteria can also affect their secretion of metabolites.
84. Emergent phenotypes involve the cross-protection of different microorganisms when undergoing sensitisation following exposure to drugs. This particular vector in the community through bio-accumulation or biodegradation, or sometimes using both mechanisms, actually protects other members of the community in the microbiome.
85. Attention was drawn to the impact of common chemical pollutants on the gut bacteria, which includes environmental contaminants, pesticides, and food contact materials. An image was presented of the pollution library which contains more than a thousand different compounds, including pesticides and industrial chemicals. This library shows the broad ranges of chemicals that can affect the human microbiota and the wide range of effects they can have on the bacteria and how this is comparable to the effects of human-intended pharmaceuticals.
86. The importance of viewing these chemicals from a biological perspective, and not only from the chemical perspective, is critical when categorising them and considering how they need to be controlled to prevent bacterial resistance, and other damage.
Potential adverse effects of pesticides on the microbiome
87. Mr Neil Lister (Crop Life International Representative) firstly introduced the WHO/FAO Joint Meeting on Pesticide Residues (JMPR) who have begun requesting available data on the impact of pesticide residues on the human gut microbiome when recommending Codex Maximum Residue Limits and proposed testing using a guideline developed for antimicrobial veterinary medicines.
88. Crop Life’s main questions are surrounding: the outstanding safety questions relating to potential effects of pesticide residues in food on the human gut microbiome and how well advanced is the scientific understanding of the human gut microbiome and what is the likelihood of pesticide residues in food, at the concentrations at which they are present, having an adverse effect?
89. The speaker then discussed what information is already available for pesticides. Each pesticide active ingredient is assessed by generating a large mammalian toxicity dataset (acute toxicity, genotoxicity, neurotoxicity, developmental and reproductive toxicity, and repeat dose systemic toxicity (short and long term)), mostly obtained using oral dose in-vivo studies. Adverse health outcomes that may be mediated by the gut microbiome have likely been assessed and incorporated into the health-based guidance values derived from these data, albeit indirectly.
90. A current data gap is specific experimental data on the impact of pesticides directly on human gut microbiomes. These datasets are rarely available (although they may exist as historical data) and are not currently required by regulatory agencies.
91. In summary, further science and development is needed to investigate how pesticides impact the human gut microbiome and its influence on human health, specifically in:
- Defining a healthy microbiome and understanding normal fluctuations in the microbial composition.
- Understanding how changes in the microbiome relate to adverse health effects.
92. Finally, the speaker prosed some questions that will need to be addressed. including what additional mammalian hazards are not addressed by current data? and if specific experiment data for a pesticide are required, a globally harmonised and validated test guideline (ideally via the OECD) should be developed.
Microbiological Risk Assessment of Residues of Veterinary Medicines
93. Dr Silvia A. Piñeiro (U.S. Food and Drug Administration (FDA)) introduced her talk which focused on how new animal drugs are assessed and the importance of their effects on the human intestinal microbiota.
94. In traditional toxicology, an assessment is performed to establish an Acceptable Daily Intake (ADI) and determine a safe concentration for veterinary drug residues in food. The ADI is an estimate of the amount of a substance, expressed on a body weight basis, that can be ingested daily over a lifetime without appreciable risk to human health.
95. For antimicrobial drugs, a human intestinal flora assessment needs to be performed to determine whether a microbiological ADI (mADI) is necessary (Piñeiro et al., 2021).
96. The concern for antimicrobial residue effects on the human intestinal microbiome is that the human intestinal microbiome is a complex group of microorganisms composed of bacteria, fungi, archaea, protozoa, and viruses, organized in a community, living in close relationship with their host and the human gastrointestinal tract environment. This is a balanced, diverse, and complex ecosystem performing essential functions that may be disrupted by drugs and their residues.
97. In vivo and in vitro systems have shown that very low levels of drug residues from edible animal tissues can alter the intestinal microbiome and may result in human health consequences.
98. Drug residues present in edible animal tissues may reach the human colon by the oral route due to incomplete absorption and may be absorbed, circulated, and excreted via bile and/or secreted through the intestinal mucosa.
99. United States developed guidance for industry (GFI) #159/VICH GL36 and first implemented the guideline in 2005. It describes the approach for establishing mADI’s.
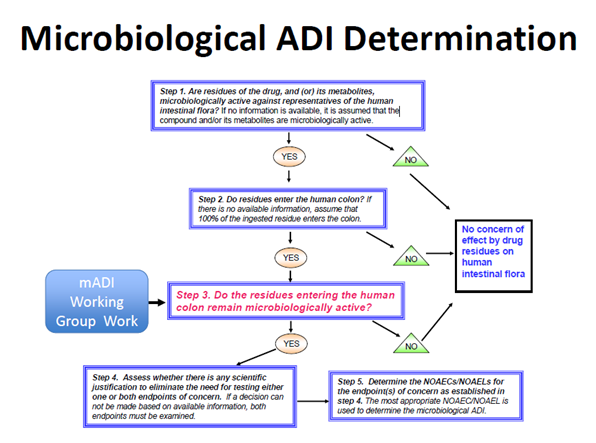
100. A five-step decision tree illustrates the approach for determining whether drug residues reaching the human colon remain microbiologically active, and whether a mADI determination would be necessary for drug approval (Figure 1).
Figure 1. Microbiological ADI Determination. Figure taken from Piñeiro, S.A. and Cerniglia, C.E., 2021. Antimicrobial drug residues in animal‐derived foods: Potential impact on the human intestinal microbiome. Journal of Veterinary Pharmacology and Therapeutics, 44(2), pp.215-222.
101. It recommends in vivo or in vitro test systems and methods for determining no-observed adverse effect concentrations/levels (NOAEC/Ls) for endpoints of human health concern and a procedure to determine a mADI from the NOAEC/Ls. The endpoints of concern are: disruption of the colonization barrier and development of antimicrobial resistance.
102. When establishing the mADI, the microbiological endpoint with the lowest value is used; either disruption of the colonisation barrier or an increase in the population(s) of resistant bacteria.
103. There are multiple approaches to addressing the impact of residues on the human intestinal flora based on Veterinary International Conference on Harmonization (VICH) GL36.
104. Six mADI examples were presented based on a variety of microbiological endpoints: tilmicosin (disruption of colonization barrier); narasin (disruption of colonization barrier); lincomycin (disruption of colonization barrier); tetracycline (Increase in the population of resistant bacteria in the human colon); amoxicillin – Joint FAO/WHO Expert Committee on Food Additives (JECFA, 2017).
105. For amoxicillin, a microbiological ADI of 0-0.002 mg/kg bw could be established on the basis of disruption of the colonization barrier of the gastrointestinal tract and using the adopted colon content volume of 500 mL. For ampicillin reviewed by JECFA in 2017, the overall mADI of 0-0.003 mg/kg bw was based on the increase in the population of ampicillin-resistant bacteria in humans, and using a safety factor of 10, as this microbiological end-point is lower than the microbiological ADI for its effects on colonization barrier disruption.
106. The GFI #159/VICH GL36 was later revised after new research and discussions.
107. The addition of Appendix D in 2013 ‘Regarding the Determination of the Fraction of Oral Dose available to Microorganisms’ was a key step forward in improving the guidelines. The change of the colon content volume to 500 mL/day, implemented in the US in 2022 was also highlighted.
108. Finally, the speaker pointed out the data gaps which include:
- what can be considered a normal microbiome?
- what are the most sensitive indicators/biomarkers of microbiome dysbiosis and how do single and combined toxic exposures effect the intestinal microbiome?
- Metagenomics vs metatranscriptomics vs metabolomics?
- What about the applications of new methodologies for establishing a mADI?
- What are sensitive indicators/biomarkers of dysbiosis?
- Changes in populations? Changes in microorganism functions (metabolomics, immunity?)
Session II Roundtable Summary
In this guide
In this guideSession II Roundtable Summary
How should we consider chemical-microbiome interactions from the two aspects: microbiome modulation of toxicity and the toxicant modulation of the microbiome?
- The ADI and the NOAEL were deemed an important and effective way of establishing what is a measurable concern of the microbiota.
- The gut microbiota may be a tool to protect a therapeutic target.
- Consider whether the effects seen are reversible and whether a toxicological end point refers to permanent damage or a temporary fluctuation.
- There was a view that the microbiome was unique for an individual; however, there could be potential trends and/or patterns to establish principles to help assess the risk. For example, correlating genetics, disease states, groups that are on certain medications to the metabolome produced by their microbiota.
- The importance of understanding the mechanism of action of a chemical was reiterated as this information is necessary to understand any health effects and would be required before meaningful risk assessment could be undertaken.
- When looking at the effects of the microbiome, it is important to also look at pre-conception, as the next generation would be exposed from stages well before conception, so would need to include information on e.g. fertility and developmental effects.
How can chemicals be tested for the effects of concern resulting from changes in the microbiome?
- Discussion arose around using animal models. Pigs seem to be a useful model organism as their microbiome reflects that of a human quite closely.
- Use of animals isn’t allowed for testing of cosmetics so therefore in vitro methodologies need to be used but there are questions on whether the hugely complex interactions that occur in the microbiome can be fully reflected by in vitro tests or indeed by in vivo tests not conducted in humans.
- With regards to the skin microbiome, questions arose on the restrictions on using animal models when studying the impact of pollutants on the skin microbiome.
- The importance of cause and effect was raised, specifically the need to understand if a change in the human microbiome was due to the action of the microbiome on a chemical or the action of the chemical on the microbiome. It was indicated that there is currently not enough information available to understand the pathology associated with fluctuations in the human microbiome.
- Simplified in vivo models can help establish baselines across populations, before establishing effects of chemicals on the microbiome
- Some attendees disagreed that the standard OECD test guidelines for in vivo toxicity studies were suitable for determining effects on the microbiome. OECD guideline studies did not reveal a difference in a microbiome population and/or functionality in a 90-day rodent study, whereas omics analyses did, and current development of these techniques is resulting in a more confident prediction of health outcomes.
Main themes
- Sensitive indicators and biomarkers of dysbiosis.
- Causation vs Correlation.
- The ADI and the NOAEL were deemed useful concepts in establishing what is an impact of concern on the microbiota.
Session III Assessing the impact microbiome
In this guide
In this guideSession III Assessing the impact microbiome
Intestinal organoids as in vitro models for assessing host-microbiome interactions and safety of intervention
109. Dr Tamas Korcsmaros (Imperial College London) introduced his talk, which focused on intestinal organoids, and their use as in vitro models to assess interactions between the host and the microbiome.
110. A chart was shown comparing the number of procedures carried out for animal research in Great Britain in 2020. Imperial College London was in the top 10 with 63,670 procedures, however it was commented that it was hoped that this value would decrease in coming years.
111. The speaker explained that organoids are ex vivo primary cultures capable of self-renewal and self-organisation and exhibited similar three-dimensional structure and functionality as the tissue of origin. The concept of this is that structures could be grown from adult cells with the functionality of organ tissues.
112. Organoids allow for the modelling of different organs, including those that cannot be obtained from biopsy such as the brain. Furthermore, organoids allow for personalised modelling including the interactions between different cells; genetically modified organoids can also be used to examine specific diseases and for omics analysis.
113. Although organoid technology for screening is still estimated to be another 3-5 years away, there are currently some established screening approaches.
114. The first of the two main approaches is the many to one approach, which uses a fixed genotype organoid exposed to various metabolites, microbial species, or other molecular libraries. The second is the one-to-many approach, in which organoids with different genetic backgrounds are exposed to the same molecular or microbial species.
115. An example of this in practice is the human Autophagy Reporter Colon Organoid (hARCO) line. It was noted that organoid technology is very expensive for both the standardisation and optimisation aspects. An example of how this technology would be relevant using gut organoids was illustrated.
116. Problems with traditional approaches were raised. It was explained that in the epithelium layer in the colon there are many cell types, however these are not included in traditional cell cultures. Additionally, a lot of knowledge currently comes from mouse models, yet the microbiomes of mice and humans are not comparable and could work differently. Organoids would eliminate these problems as all cells would be included and these would be grown from humans.
117. Issues arise, however, when growing intestinal organoids as the luminal side, which would be the outside layer in the gut, becomes the inside layer in the organoid model. Although this still allows for the study of epithelial homeostasis, regeneration, cell-cell interactions and intracellular processes, it is not appropriate to study cross microbe interactions.
118. To overcome the difficulties in this technique, micro-injections can be used to administer microbes to the middle of the organoid to interact with the lumen, or an easier but less efficient method would be to flip the organoid so that the basolateral side is inside and the lumen outside. Work is also undergoing to create a 2D system with both a luminal and basolateral side.
119. It was explained that there were multiple models currently available, all varying in complexity, yet the aim was to be able to produce a ‘Gut-On-A-Chip’ microfluidics systems. Work towards this is now being set up in Imperial College London in collaboration with a number of companies. These chips would work by combining organoid cells, patient metadata, and microbiota and nutrients, which would be characterised and undergo multi-omics analysis to allow use of these systems for screening.
120. It is hoped over time these systems will allow for the study of cell-cell and cell-microbe interactions and be useful diagnostic and prognostic tools in combination with omics approaches.
Analytical strategies to study the gut microbiome in toxicology
121.Professor Michael Antoniou (Kings College London) introduced the topic by outlining the parameters required for microbiota compositional and metabolic function investigations. These included:
- Determination of both bacterial and fungal populations.
- Gut omics analysis (transcriptomics/proteomics/metabolomics).
- Gut integrity measures.
- Correlations with internal organ/system analysis.
- Functional studies in vitro that can complement in vivo investigations.
122. The speaker then presented the results of three studies that highlighted the effects of glyphosate-based herbicides on gut structure and function. These herbicides are non-selective (i.e. broad spectrum), are the most heavily applied globally and their worldwide spread of use and usage continues to rise.
123. It was highlighted that commercial glyphosate-based herbicide formulations contain many additives (co-formulants/adjuvants) in addition to glyphosate with the co-formulants shown to be toxic in their own right. Thus, it was emphasised that toxicity studies whenever possible need to compare glyphosate with typical commercial formulations, since the latter can be far more toxic than glyphosate alone.
124. The mechanism of action of glyphosate was introduced. In brief, it interferes with the shikimate pathway in plants and thus inhibits formation of aromatic amino acids. It was once thought that this pathway was exclusive to plants, however, it is also present in some bacteria and fungi, including those in the gut of animals and humans.
125. The first results presented were from a comparative toxicogenomics study of glyphosate and typical EU glyphosate-based herbicides using an in vitro murine embryonic stem-cell based genotoxicity assay and in vivo molecular profiling (omics) in Sprague-Dawley rats (Mesnage R et al., 2021). Marked metabolic disturbances in the gut in both treatment groups were observed even though there was little change in the rat’s gut microbiome composition. The metabolic changes were reflective of the treatments inducing oxidative stress.
126. The second results presented were from the Global Glyphosate Study, focusing on the effects of prenatal exposure to glyphosate, 2,4-D and dicamba, when in the formulation on gut function and integrity in Wistar rats. Both treatment groups decreased bacterial diversity and increased fungal diversity in the gut.
127. Data from unpublished work was also presented where Wistar rats starting at prenatal stage of development were treated with either glyphosate alone or as a mixture with two other highly used herbicides in the USA, 2,4-D and dicamba. Glyphosate at the UK/EU no-observed adverse effect level dose and more so the mixture of glyphosate/2,4-D/dicamba at each at the UK/EU acceptable daily intakes caused alterations in gut bacterial and fungal composition, inflammation, redox imbalance and compromised integrity (“leaky gut”).
128. Also presented was a computational study drawing on data from the human gut microbiome database. Among the 44 subspecies reference genomes, (72% of the total assigned microbial abundance in 2144 human faecal metagenomes), 35 species are predicted to be sensitive to glyphosate. Thus, it was shown that glyphosate can potentially affect the human gut microbiome (Mesnage R & Antoniou MN, 2020).
129. The final results presented were from a study conducted to evaluate the effects of glyphosate and a typical US Roundup commercial formulation on the gut microbiota of a healthy 3-year-old child using the SHIME® (Simulator of the Human Intestinal Microbial Ecosystem) technology (Mesnage R et al., 2022). It was observed that Roundup and to a lesser extent glyphosate caused changes in fermentation and metabolic activity: i) increased lactate and acetate caused acidification of the microbiological environment; ii) decreased short chain fatty acids and iii) increased long chain polyunsaturated fatty acids. It was also found that Roundup increased ammonium production reflecting increased proteolytic activity.
130. To conclude, these studies showed that several analytical methods are required to holistically evaluate the potential health effects of glyphosate and its commercial formulations on the human microbiome. This included omics, biochemical/gene expression and histological measures, molecular profiling analyses and the SHIME® system.
131.Several regulatory recommendations were put forward for consideration.
These were:
- Multi-omics analyses should become an integral part of chemical toxicity evaluation.
- Chemical administration in vivo should begin pre-natally and preferably continue life-long to more accurately reflect real world exposure scenarios.
- Gut and internal organs/systems need to be assessed in parallel.
- Long-term toxicity testing of commercial pesticide formulations as well as active ingredients is needed.
- Pesticide risk assessment and acceptable daily intake values need to be established based on tests of chemical mixtures.
Session III Possible ways to evaluate in the short to medium term and microbiome interventions for maintaining health and treating disease
In this guide
In this guideSession III Possible ways to evaluate in the short to medium term and microbiome interventions for maintaining health
and treating disease
A tiered approach to risk assess microbiome perturbations: illustrative case study on effects induced by application of beauty and personal care products
132. Dr Aline Métris (Unilever) introduced a tiered approach to risk assess the effects of beauty and personal care products on the skin and oral microbiome (Métris et al., 2022). The presentation was focussed on the skin microbiome and began by discussing the large biogeographical variability (i.e. elbows different from hands which are different from face) as well as significant inter-individual variation in the microbiome.
133. Although several dermatological conditions (including atopic dermatitis) are associated with dysbiosis of the skin microbiome, it is uncertain what precisely characterises a ‘healthy’ skin microbiome. A similar theme is reflected across many discussions on the microbiome and hence, at present, a relative, rather than an absolute, approach to risk assessment is more feasible. This is where an individual serves as their own control, whether in time or space.
134. With these considerations in mind, the speaker then discussed the Unilever Safety, Environmental and Regulatory Science (SERS, previously SEAC) Microbiology Team approach to tiered risk assessment (Figure 2) in the context of the skin microbiome.
135. A tiered approach is one in which evidence of risk at one level (or tier) prompts progression to the next level (or tier), as necessary, and in which each level is increasingly empirical and experimental in nature.
Figure 2. Tiered framework to assess the risk in response to the perturbation of the oral or skin microbiome by application of beauty and personal care products. The risk is classified into “low risk” (green boxes) and “uncertain risk” (orange box). The blue boxes refer to the first tier which is based on the notion of History of Safe Use, the second tier, depicted in turquoise, is based on the notion of reversibility of effect on key species and the third tier, in red, relies on Next Generation Sequencing (NGS) data describing taxa/functions. The data type used in each tier is shown under the corresponding box. Figure taken from Métris et al., 2022.
136. Questions such as ‘is the product a direct or indirect antimicrobial’ should guide whether an assessment is required at all. Tier 1 is based on evidence of history of safe use of a specific compound or other technologies that have the same or more pronounced effects on the microbiome than the compound under assessment. In the absence of history of safe use, one needs to progress to Tier 2.
137. Tier 2 is based on the reversibility of any change induced by the personal care product. In a longitudinal study, the microbiome is compared between baseline (i.e., before product application), after product application (intervention period) and after discontinuing product use (regression). After stopping using the product, do any observed changes during intervention return to baseline? Important considerations include the precise timing of the protocol (i.e., the length of intervention and regression time) and the methods used to profile the microbiome. Methods include 16S ribosomal RNA (rRNA) sequencing for microbiome profiling and Quantitative PCR (qPCR) for quantitative estimates of species of interest. However, functional changes and compositional changes may be related in relatively complex ways, meaning that this step requires careful analysis. If the observed change is not reversible then it is necessary to proceed to Tier 3.
138. Tier 3 involves an analysis of the microbiome functions implicated in the change and an understanding of which mechanisms are important for maintaining skin and oral health.
139. Finally, the speaker summarised the remaining scientific challenges by asking the questions:
- Which organisms/strains/species are biomarkers of health and disease?
- Which are the corresponding functions impacted and what might the effects be?
140. Known microbiome functions include resistance to colonisation of pathogens and interactions with host barrier functions and immunology. However, the specific modes of actions are not well understood. In silico and in vitro methods may be adopted to characterise microbiome functions (rather than taxa) and host-microbiome interactions.
Faecal Microbiota Transplant (FMT) Liver disease example
141. Dr Lindsey Ann Edwards (Kings College London) presented on how the microbiome can be modified using faecal microbiota transplantation in patients with end-stage cirrhosis.
142. The focus of the research is on modulating the microbiome to reduce intestinal barrier damage, inflammation and antimicrobial resistance. Simultaneously, reducing infections in chronic inflammatory diseases, including liver disease, lowers the requirement for the prescription for antibiotics.
143. Globally, 8 million people per year die due to infections; of those, over a million of these deaths are due to drug-resistant organisms, resistant to all known antibiotics. Antimicrobial resistance is escalating, and no new antibiotics have been discovered since the 1980’s. Therefore, the Lord O’Neil report predicts deaths due to antimicrobial resistance could reach over 50 million per year by 2050 if urgent action is not taken.
144. Liver cirrhosis patients (end-stage liver disease) have a microbiome dysbiosis leading to an outgrowth of pathogenic species that causes intestinal barrier damage and inflammation, and translocation of microbial species across the intestinal barrier, which can lead to systemic infections. This is combined with a disrupted immune system that is not able to fight those infections, which then leads to multiple organ failure. A dysbiotic microbiome causes intestinal inflammation, which can act as an environmental stressor to the microbes, which allows for the upregulation of virulence factors such as antimicrobial resistance genes, which progresses as the liver disease progresses. This increased risk of infections, particularly drug-resistant infections, is a major cause of deaths in patients with liver disease.
145. The microbiome within the intestine interacts with a single epithelial layer and a large mucosal immune system. In fact, 75% of the body's immune system is in the intestine. The epithelial layer and the mucosal immune system are responsible for preventing microbial translocation. Approximately 75% of the blood from the gut drains to the liver. If microbial translocation occurs, then the microbes can end up in the liver where there is also an immune system to try to tackle the infectious agents. This is the body’s final resort in stopping the systemic spread of infection and possible septicemia and death.
146. As liver disease patients are prone to risk of infection, they are prescribed prophylactic antibiotics even when no infection is detected. Many liver patients take them daily whether needed or not. This practice can exacerbate the situation as the antibiotic also acts as an environmental stressor and can lead to further antimicrobial resistance. The ATTIRE trial was published in the New England Journal of Medicine, finding that in the absence of an infection, there was ‘No evidence’ that prophylactic antibiotics prevent hospital-acquired infections in cirrhosis patients and recommended targeted antibiotic prescription on the development of an infection. However, as the risk of infection and death is high in end-stage liver disease, it is understandable that many liver clinicians are reluctant to cease the prophylactic prescription. Clear guidelines are required so this practice does not continue.
147. The NIHR-funded PROFIT study was a randomized placebo-controlled trial of Feacal microbiota transplant (FMT) in liver cirrhosis; the aim is to use a treatment that modifies the microbiome in order to reduce inflammation and the spread of antimicrobial resistance, as well as treat the cirrhosis. Donor stool was screened to determine no carriage of pathogens and was blended with glycerol, then administered into the duodenum via a naso-gastrointestinal tube inserted during endoscopy.
148. Exclusion criteria were that the recruited patients were not consuming alcohol and were not on any antibiotics. The recipients were mostly males in their 60s who had consumed diets high in animal protein and sugar, the donors were females in their 20-30s who followed either a vegan, vegetarian or omnivorous diet high in fruit and vegetables and complex carbohydrates.
149. Stool samples were taken at baseline before treatment and then 7-, 30- and 90-days post-treatment. Various techniques were employed to examine the effect of FMT on the microbiome itself and any changes in patient immunity and the intestinal barrier were studied, including plasma and faecal cytokines and intestinal barrier integrity markers of the gut. It was shown that FMT allowed for donor engraftment which is important to success.
150. It was shown that a mixture of a high-fibre, high-protein diet with FMT allowed for a more omnivorous microbiome. After one dose of the FMT, species richness increased from the baseline within the recipient; this did start to drop off at around 90 days, but did not return to the baseline level. The results suggested that multiple FMT doses would likely be needed. The new NIHR-funded PROMISE trial is dosing patients every 90 days.
151. Dr Edwards has discovered that FMT reduced pathogens, including those that cause epithelial barrier disruption, which are more prevalent in those that consume a western diet. This occurred in combination with the replacement by many anaerobic beneficial species of those consistent with a diet high in complex carbohydrate consumption, where microbial metabolic capacity was also replaced.
152. Dr Edwards has developed a method that allows for the quantification of mucosal cytokines by electrochemiluminescence. T helper 17 cell (TH17) cytokines and markers of barrier disruption were measured using this technique. It was found that the patients had levels of mucosal TH17 cytokines sufficiently high to damage the intestinal barrier, with low levels in the circulation, reducing the ability to fight infections. Treatment with FMT facilitates a reversal, which enables barrier repair and to fight against infections.
153. A mass-spectroscopy technique was developed by Dr Edwards to identify faecal proteins. Analysed pre- and post-FMT, in comparison to placebo controls, FMT resulted in changes in the levels of a number of proteins, of which 301 were quantified, 154 of human origin, and 147 bacterial. The human proteins were those involved in barrier response and repair. There was also evidence of a change in xenobiotic metabolism. However, this will require further research. Most of the bacterial-derived proteins were enzymes, including those involved in ammonia metabolism. In liver disease, the liver is unable to metabolise ammonia, therefore, the ammonia is retained in the blood and leads to hepatic encephalopathy, which can be fatal.
154. After FMT, a reduction in plasma ammonia is seen and there is an increase in faecal ammonia achieved by a change in metabolic pathways, which in turn leads to a better functioning of host immunity. In the future, boosting better immunity may reduce deadly infections and may limit the need for antibiotic prescriptions.
155. Dr Edwards found that faecal microbiota transplantation also reduced antimicrobial resistance gene carriage.
Session III Roundtable Summary
In this guide
In this guideSession III Roundtable Summary
How does the microbiome impact on the ingredients we consume?
- The potential exists that a change in the microbiome can have negative effects on drug metabolism and therefore affect the bioavailability of drugs. This raised an interesting point from the reverse perspective on how we protect the microbiome to be responsive to xenobiotics and not impact their mode of action for treating problems elsewhere in the body.
What are the human health outcomes of concern that are related to gut microbiome-mediated change, and what do we know about the relative sensitivity of risk assessment of other toxicological endpoints in response to these effects?
- Patients that are potentially suffering from liver or kidney disease could have increases in sensitivity to changes in the microbiome, producing higher concentrations of different molecules that could exacerbate disease.
- An example was given of ketones being produced by bacteria and the high susceptibility of those with kidney disease.
- The issue of mental health-related effects associated with microbiome changes is an area that needs to be prioritised further.
- How to define adverse effects of changes in the microbiome, or measure impact, is not going to be easily defined.
- Humans have access to dietary protective factors, e.g. anti-inflammatory and high antioxidant diets, but consumers are also exposed to unhealthy, poor diets.
- In terms of risk assessment, to account for the possibility of microbiome variation and microbiome-mediated toxicity, consideration could be given to covering this by the application of an additional uncertainty factor.
- The effects of the microbiome on toxicokinetics, including enterohepatic recirculation and metabolism of xenobiotics into toxic metabolites were noted.
Main themes
- Accounting for microbiome variation and microbiome-mediated toxicity could be achieved by an additional uncertainty factor.
- Effects of the microbiome on toxicokinetics, including metabolism of xenobiotics into toxic metabolites should be considered.
Session IV Future Directions
In this guide
In this guideSession IV Future Directions
Biotechnology and Biological Sciences Research Council (BBSRC)
Integrative microbiome research and capability
156. Dr Louisa Jenkin (BBSRC) presented on BBSRC’s interests in microbiome research and what their current activities in this area are.
157. The UK Research and Innovation (UKRI) invest £8 billion per year in research and innovation partnering with academia and industry. The BBSRC support bioscience research through training and investments (£378m).
158. BBSRC interests in microbiome research are to understand the biological mechanisms of the role and function of the microbiome on health.
159. The microbiome fits under the remit of the BBSRC in their goal to improve food safety by better understanding the risks to human health from food at any stage of the food chain. This also involves understanding the unintended consequences of food innovation, processing and dietary change, which includes the microbiome.
160. The BBSRC does not conduct research focused on specific human diseases or abnormal conditions; animal models of human disease and human toxicology; or human-human transmission.
161. BBSRC held a workshop in 2020 on Microbiome Capability in which 45 participants attended from 38 research organisations, industry, networks and societies. The aim was to help the BBSRC to understand current and future microbiome community needs.
162. The speaker described the general reflections of the workshop, the biggest research challenges identified by the participants and what the UK’s capabilities in the area are. There is a need to build interdisciplinary approaches, and collaborations will be crucial, to understand the functions of microbiomes and the influence of key factors. It will be important to connect basic and applied research; look at all components of the microbiome; increase the variety of model organisms/ choose the right model system, including the use of synthetic communities to explore complex interactions; share data, software, tools and methodology within and between communities and fields as well as the integration of data, e.g. multi-omics; data standards, metadata, Findable, Accessible, Interoperable, and Reusable (FAIR).
163. Some of the biggest research challenges for the microbiome field include correlation and causation; sampling; scales, time series and longitudinal studies and functional annotation.
164. It was noted that the UK current technologies, resources and skills in the field include data and computing; infrastructure; multidisciplinary collaboration skills and expertise; as well as translation.
165. The workshop helped to inform new BBSRC activities: BBSRC-NSF/BIO lead agency 2024 and BBSRC-DFG Lead Agency Pilot 2023-2024. These aim to help understand host-microbe interactions, synthetic microbial communities and the microbiome as a whole. It also fed into other activities such as the KTN Microbiome Innovation Network road-mapping activity and strategy.
166. The speaker then stated some of BBSRC strategic investments and current activities including: the Quadram Institute, which is focusing on understanding how gut health, microbiology and food interact to promote physical and mental health across the life course and prevent disease including microbial safety in the food chain; and the Centre for Microbial Interactions to harness the huge breadth and capacity of our expertise within the extensive microbiology community at Norwich Research Park (NRP) to solve global challenges in food security, human health and climate change (which includes chemical interactions: AMR, natural products cell biology).
167. In Summer 2022, co-funded with the Food Standards Agency, a £2.2m Innovation Hub for Food Safety and UK Food Safety Research Network were launched to coordinate and fund cross-sectoral research and training activities that address current and emerging challenges in food safety. The aims include addressing microbial risk in the food chain, with the goal of introducing new capability, knowledge or skills to help reduce these risks.
168. BBSRC has recently announced £378m to support 16 new programmes, which cover a wide breadth of bioscience research. The programmes cover areas such as advanced genomics, crop resilience, animal health & welfare, food safety, food security, nutrition, lifelong health, preventing & controlling viral disease, and sustainable farming & agriculture.
169. The research themes include a variety of microbiome work: chemical interactions (antimicrobial resistance (AMR)), engineering microbial interactions (synthetic biology, biotechnology), microbial community interactions (microbiomes, infection, health), plant-microbe interactions (mutualism, symbiosis, pathogenesis), evolutionary interactions (biodiversity, genetics, genomics).
170. Finally, the speaker presented relevant BBSRC investments into microbiome related work encompassing biofilms, AMR, infectious diseases, diet and health, gut immunology and the skin microbiome in healthy aging.
171. Moving forward, BBSRC is taking a one health approach to improve interdisciplinary working and the interconnectedness of soil, plant, animal and human health, considered alongside the complexities of wider health, social and environmental factors to avoid adverse effects, unintended consequences and promote nutrition security.
European Food Safety Authority (EFSA) Roadmap Microbiome
172. Dr Javier Moreno (Food science Research Institute (CIAL)/Spanish National Research Council (CSIC)) introduced the EFSA roadmap, which includes the impact of the microbiome on human, domestic animal, and plant health.
173. This project has performed a comprehensive and critical assessment of the evidence-based research about: i) the impact of dietary compounds on the human and some domestic animals (i.e., poultry, ruminants and pigs) gut microbiome; ii) the most representative in vitro and in vivo models of the human gut microbiota currently used in microbiome research studies; and iii) the methodology used to measure changes in the microbiota.
174. The approach used was to critically review the knowledge, data, and methods relevant for risk assessments. This, together with a number of experimental case studies, which looked at impacts on human and animal health, enabled identification of gaps in both the literature and research.
175. The draft proposed roadmap aims to advance research for addressing risk assessment needs, accounting for effects on/by the microbiome in humans, domestic animals, and the environment.
176. Drafting a roadmap for the gut microbiome is a real challenge. The gut microbiome plays an important role as a mediator and moderator of the effects of some dietary compounds on humans/animals. Incorporating these gut microbiome interactions in the risk assessment could help to fully understand potential hazards derived from their exposure.
177. The gut microbiome is a complex universe by itself, even more so when looking at its relationship with the host. It is a relatively novel research area that is evolving in parallel to technological developments and bioinformatics.
178. Knowledge gaps include: i) lack of data on the exposure of the gut microbiome to certain compounds and underlying mechanism(s); ii) lack of standardized models, iii) methodological limitations, iv) lack of guidance to evaluate microbiome-related data in safety evaluations.
179. The first part of the project was to establish how the gut microbiome acts as a moderator for dietary compounds in humans and animals. This needs to be inclusive of dietary compounds that are partially or not absorbed and incorporate the potential hazards of this into any further risk assessments.
180. The stages of the roadmap (Figure 3) comprise the following steps:
1) Prioritisation of compounds to be assessed
2) Research strategy
3) Policy action- Decision framework.
Figure 3. Roadmap to advance research to address risk assessment needs to account for effects on/by gut microbiomes in humans. Figure taken from EFSA Roadmap for the integration of gastro-intestinal (GI) tract microbiomes (human and domestic animal) in risk assessments under EFSA's remit (2024).
181. The speaker then discussed the interactions between dietary modulators, the gut microbiome and the host (Garrido-Romero et al., 2024). There is evidence of some fatalities from chemical contaminants in food potentially due to detrimental effects on the human gut microbiome. These studies have been focused on the understanding of the mechanisms, which include:
- The gut microbiome can metabolise xenobiotics directly through microbial enzymatic biotransformation by specific bacterial enzymes. This could lead to the production of toxic metabolites that can then be absorbed in the small intestine and/or liver where they are detoxified and form conjugates. These are then secreted back into the intestinal lumen and into the microbiome community where the active toxic moiety is released, which can potentially cause carcinogenic or genotoxic effects.
- Another mechanism of xenobiotics is an indirect influence on host metabolic and transport pathways in the liver utilising microbial products.
- The inhibition or promotion of bacterial growth which can affect the composition and function of the gut microbiome and disrupt the homeostasis of the environment.
182. The speaker then emphasised that there is a need for internationally agreed minimal methodological requirements and reference materials for research and regulatory bodies to use when analysing the human gut microbiome. This needs to include information on sample collection, storage/processing of samples, handling and generating data, analysis workflows and fit-for-purpose data repositories for risk assessment.
183. There is a need to develop fit-for-purpose translational models to identify potential key events following exposure to harmful diet-derived compounds and predict the potential effects that can be had on the gut. Fit-for-purpose translational models need to be a considered as validated experimental models, because this is essential in the overall context of assessing effects on the gut microbiome. The gut microbiome can be a critical player in the metabolic and inflammation processes, immune reactions, and integrity of the gut barrier. Validated models can have short to medium exposure endpoints set as the markers for experiments. This can be using inflammatory markers, which assess the integrity of the epithelium, combined with markers produced by the bacteria that signal disruption.
184. Finally, the speaker discussed risk assessment and policy actions. Policy actions are included in the roadmap to help with regulator and innovator engagement to address these gaps in knowledge, and also to promote and prioritise funding areas for research and regulatory needs. The Roadmap encourages collaborative efforts to develop decision trees and end points that can be used by risk assessors and policy makers, enabling both safety and efficacy assessments related to microbiome-based products and tools. In particular, this will help close the gaps in early-microbiome regulatory guidance and ensure that it is fit for purpose in practice.
Microbiome Strategic Roadmap (Regulatory and food)
185. Professor O’Brien (The Food Observatory and Ulster University) introduced the KTN Microbiome Special Interest Group (part of Innovate UK) to the audience. The objective is to bring together all stakeholders with an interest in the microbiome and to increase the translation of knowledge on the microbiome from science into action. Representatives of the food and pharmaceuticals industries, regulatory bodies, research funding councils and academia form this group, which has now been renamed as the Microbiome Innovation Advisory Board.
186. Two conferences organised by the Microbiome Innovation Advisory Board have already been held, one in Glasgow in 2023 and another one in Liverpool in 2024. The speaker explained that the future of this board is to hand the responsibility of delivering the recommendations for a microbiome strategy roadmap to a multi-university consortium group.
187. So far, three reports have been published by the Microbiome Innovation Advisory Board:
1) Microbiome Strategic roadmap
2) Human Intestinal Microbiome Therapies and Diagnostics – The Science, Opportunities and Challenges
3) Securing the future of microbiome research and innovation: the need for biobanking infrastructure in the UK
188. The presenter noted that the KTN Microbiome Strategic Roadmap is in line with the One Health approach to support unifying research activities in human, animal and plant health, and hopes to advance science translation and business application. The scope of this roadmap encompasses a wide range of applications, e.g. human intestinal and plant microbiome; animal, plant, crop and soil health; and live biotherapeutic products.
189. Within the nutrition and wellness areas, the speaker enumerated a few sub-areas such as immune modulation and foodborne diseases, and the most research driven areas i.e. metabolic health, women’s health and ageing.
190. The presenter went on to highlight that one of the goals of the KTN Microbiome Strategic Roadmap from a regulatory perspective is to anticipate and mitigate any unintended consequences of a product, to be ahead of any issues that might give raise to future human health concerns. The goal is “Ensuring regulations, rules and good regulatory practices encourage advances that target unmet needs, mitigate any unintended consequences of the developments and are based on good regulatory principles”. This goal is shared with the National Science and Technology Framework and National Vision for Engineering Biology.
191. The speaker then stated that the UK is well positioned to take advantage of opportunities represented by microbiome therapeutics and diagnostics. However, they also highlighted some of the challenges of this roadmap. For example, there is a gap between innovation and regulatory procedures, and also a lack of regulatory standards and definitions, which brings uncertainty. Another challenge is that assets resulting from microbiome research can impact multiple regulatory domains, e.g. food, agriculture, animal health, aquaculture, personal care. Additionally, the KTN Microbiome Strategic Roadmap could potentially disrupt traditional assumptions about definitions of health and disease.
192. The presenter reminded the audience of those methodological challenges and limitations when assessing the microbiome that had been highlighted throughout the session on numerous occasions. For example, a study that had used a bacterial fingerprint to predict vulnerability to cancer had misidentified bacterial sequences with human sequences. Another example was the reported major errors in a paper that identified cancers based on their microbiome, which led to the authors retracting their article. To address these challenges, improved biological standardisation in the omics area is needed.
193. The presenter went on to introduce the topic of Adverse Outcome Pathways (AOPs). He highlighted this with a proposed AOP that does not arise from a xenobiotic, but from a SARS viral protein that interacts with enterocyte ACE2 receptors, which leads to an alteration of the microbiota. In the areas of evidence used to evaluate this outcome, one is microbiome biodiversity. At least 17 different measures are cited by the paper, however there is no reference as to what constitutes an adverse effect.
194. As a concluding remark, the speaker acknowledged that there is a significantly high variability in the human microbiome and multiple modulating factors, not just the diet. In a way, every home has its own microbiome, and that is why it is difficult to define what is normal and abnormal, what is a result of adaptation, and whether this adaptation is reversible or adverse.
Session IV Roundtable Summary
In this guide
In this guideSession IV Roundtable Summary
What are the biggest research challenges for the microbiome field?
- The main concerns and challenges are centred around the limitations on conducting mechanistic work on each bacterial species present in the microbiome. Due to the complexity of all of the different bacterial species present, defining these species and their behaviour, metabolism and contribution to the microbiome environment will be important in carrying out further research.
- Another area of importance is to establish the endpoints for defining what constitutes an adverse effect from a microbiological and/or toxicological perspective. Establishment of these will give rise to more complex questions and interactions to be studied.
- From a regulatory perspective, an increase in the available scientific literature will affect how the safety of compounds and products is evaluated. These developments will have to be considered and will require a whole new array of expertise to evaluate safety.
- Trying to distinguish between causality, correlation or association.
- Defining standard parameters for a healthy and a pathogenic microbiome.
- Infrastructure: There is need for a stool biobank in which samples could be taken at the same time as blood samples.
- Standardisation of methods so that FMT results could be compared between different research groups. The results of sequencing in one laboratory should be the same as those in another laboratory so that the data can be compared.
What are the potential future innovative and disruptive ways of tackling microbiome function and host interaction?
- Members considered the rapidly developing use of AI in research and how an increase in databases with microbiological, metabolome and genomic data will utilise AI to process information and extract relevant trends or results. This will also go hand in hand with the expansion of AI use in research and industry, which needs to be harnessed, but in a controlled manner.
- After discussing a study presented in one of the talks about how the microbiome influence is greatest at < 3 years old, members considered what is the most significant variable in the composition of the microbiome. In particular, how much can diet, and external xenobiotics impact the composition of the microbiome after its initial establishment in early childhood. If they can have a significant impact, which substances or factors have the most dramatic effect and is this what needs to be prioritised or do all potential factors have equal importance.
- Important to keep developing integrative multi-omics approaches to provide comprehensive and holistic understanding of host microbiome interaction.
- Advances in computational tools that could model the gut microbiome.
What are the current technologies, resources and skills in the field and the UK’s capabilities and capacity?
- There is limited access to laboratories and institutes that can accommodate aseptic technique studies and the use of human models in the same lab or institute.
- The importance of variety in animal and human models.
- Limitation of experts that are both computationally and lab-based trained as this is a crucial skill set to be able to interpret the kind of data produced in microbiome/metabolome-based research.
- Members highlighted the need for more AOPs to strengthen the evidence base and speculated that it is possible that a deviation in the microbiome may not be the adverse outcome in an AOP but rather one of the key events. Identifying the endpoints with which dysbiosis is associated is an important task for establishing AOPs.
What is the level of awareness and training available in the UK in this field?
- More training is needed for cross cutting issues.
- Better communication of the work being done across regulatory bodies and research.
- Take advantage of the national centres e.g. Quadram Institute.
- Still limited data, which cannot be used specifically, when conducting risk assessments, due to the absence of any concrete scientific advice or published guidance from the Scientific Advisory Committees. There was particular emphasis from COT members that it would be critical to develop UK guidance to inform the public, scientific community and regulated community on how to consider the microbiome and its impact on human health.
Main themes
- Challenge in trying to distinguish between causality, correlation or association.
- Defining standard parameters for a healthy and a pathogenic microbiome.
- Infrastructure.
- Developing integrative multi-omics approaches can provide comprehensive and holistic understanding of host microbiome interaction.
- Increase in databases with microbiological, metabolome and genomic data will utilise AI to process information and extract relevant trends or results.
Concluding thoughts
In this guide
In this guideConcluding thoughts
Microbiome is highly complex and varied between individuals. We are not yet able to define a “healthy” microbiome i.e. a baseline. Going forward we should perhaps try to define “reference populations”.
Guidelines on specification, or an ‘average’ characterisation including parameters, ranges, diversity and species information. Defining a ‘range’ of microbiomes e.g. structure, function might help identify what types of microbiomes increase the risk of adverse effects.
Investigate how microorganisms process chemicals, considering the chemical conversions that occur in the gut including toxicokinetics e.g. metabolism of xenobiotics into toxic metabolites and how this might be considered in an assessment.
Continue the development of integrative multi-omics approaches, to provide comprehensive and holistic understanding of host microbiome interactions. Functional studies in vitro can complement in vivo investigations.
Challenge in trying to distinguish between causality, correlation or association.
Increase in databases with microbiological, metabolome and genomic data, where AI can be utilised to process information and extract relevant trends or results.
Prioritisation of knowledge gaps and moving forward
In this guide
In this guideUnderstanding which changes in the microbiome relate to adverse outcomes and the extent to which these are generalizable to different sub-populations
Standardised methods for measuring microbiomes and fit-for purpose translational relevant models, which could include integrative multi-omics approaches. Explore sensitive indicators and biomarkers.
Xenobiotic chemical conversions in the microbiome should continue to be researched and how in turn they might cause adverse effects.
Guidance to evaluate microbiome-related data in chemical risk assessment evaluations.
Improve gap between innovation and regulatory procedures as well as define regulatory standards.
Use trend analysis using new methodologies to help distinguish between causality, correlation and association.
Explore fungal and viral microbiota in both environmental and human health fields not just bacteria microbiota data.
Public engagement on use of live microorganisms e.g. probiotics promoted with claims that they provide health benefits when consumed, generally by improving or restoring the gut microbiota. This can include vigilance on bacteria-host interactions and recording unwanted side effects as well as communication on the knowledge of these.
References
In this guide
In this guideReferences
Committee on Toxicity Statement on interactions between xenobiotics and the human microbiota and their potential toxicological implications: Statement on interactions between xenobiotics and the human microbiota and their potential toxicological implications.pdf
BBSRC Microbiome Capability Workshop Report (2020): BBSRC microbiome capability workshop 2020 report – UKRI
Government Office for Science: Rapid_Project_-_Microbiome_manipulation.pdf
Branchu, P., Bawn, M. and Kingsley, R.A., 2018. Genome variation and molecular epidemiology of Salmonella enterica serovar Typhimurium pathovariants. Infection and immunity, 86(8), pp.10-1128.
EFSA Roadmap for the integration of gastro‐intestinal (GI) tract microbiomes (human and domestic animal) in risk assessments under EFSA's remit: Roadmap for the integration of gastro‐intestinal (GI) tract microbiomes (human and domestic animal) in risk assessments under EFSA's remit | EFSA
FDA Guidance Document- CVM GFI #159 (VICH GL36) Studies to Evaluate the Safety of Residues of Veterinary Drugs in Human Food: General Approach to Establish a Microbiological ADI: CVM GFI #159 (VICH GL36) Studies to Evaluate the Safety of Residues of Veterinary Drugs in Human Food: General Approach to Establish a Microbiological ADI | FDA
UKRI KTN Innovate Microbiome Strategic Roadmap: Microbiome_Strategic_Roadmap_FINAL.pdf
KTN Microbiome Special Interest Group (part of Innovate UK): Microbiome - Innovate UK Business Connect
UKRI Innovate Human Intestinal Microbiome Therapies and Diagnostics – The Science, Opportunities and Challenges Report: 0489_KTN_HIMDD_Final2_AW_Updated-230228.pdf
UK Food Safety Research Network: Home - Food Safety Research Network
Garrido-Romero, M., Pazos, F., Sánchez-Martínez, E., Benito, C., Gómez-Ruiz, J.Á., Borrego-Yaniz, G., Bowes, C., Broll, H., Caminero, A., Caro, E. and Chagoyen, M., 2024. Relevance of gut microbiome research in food safety assessment. Gut microbes, 16(1), p.2410476.
James, D., Poveda, C., Walton, G.E., Elmore, J.S., Linden, B., Gibson, J., Griffin, B.A., Robertson, M.D. and Lewis, M.C., 2024. Do high-protein diets have the potential to reduce gut barrier function in a sex-dependent manner?. European Journal of Nutrition, pp.1-20.
Leeming, E.R., Johnson, A.J., Spector, T.D. and Le Roy, C.I., 2019. Effect of diet on the gut microbiota: rethinking intervention duration. Nutrients, 11(12), p.2862.
Maier, L., Pruteanu, M., Kuhn, M., Zeller, G., Telzerow, A., Anderson, E.E., Brochado, A.R., Fernandez, K.C., Dose, H., Mori, H. and Patil, K.R., 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature, 555(7698), pp.623-628.
Métris, A., Barrett, P., Price, L., Klamert, S. and Fernandez-Piquer, J., 2022. A tiered approach to risk assess microbiome perturbations induced by application of beauty and personal care products. Microbial Risk Analysis, 20, p.100188.
Mesnage, R., Calatayud, M., Duysburgh, C., Marzorati, M. and Antoniou, M.N., 2022. Alterations in infant gut microbiome composition and metabolism after exposure to glyphosate and Roundup and/or a spore-based formulation using the SHIME technology. Gut Microbiome, 3, p.e6.
Mesnage, R. and Antoniou, M.N., 2020. Computational modelling provides insight into the effects of glyphosate on the shikimate pathway in the human gut microbiome. Current research in toxicology, 1, pp.25-33.
Mesnage, R., Teixeira, M., Mandrioli, D., Falcioni, L., Ibragim, M., Ducarmon, Q.R., Zitting, R.D., Amiel, C., Panoff, J.M., Bourne, E. and Savage, E., 2021. Multi-omics phenotyping of the gut-liver axis reveals metabolic perturbations from a low-dose pesticide mixture in rats. Communications Biology, 4(1), p.471.
National Vision for Engineering Biology: National vision for engineering biology - GOV.UK
Nagata, N., Nishijima, S., Miyoshi-Akiyama, T., Kojima, Y., Kimura, M., Aoki, R., Ohsugi, M., Ueki, K., Miki, K., Iwata, E. and Hayakawa, K., 2022. Population-level metagenomics uncovers distinct effects of multiple medications on the human gut microbiome. Gastroenterology, 163(4), pp.1038-1052.
Oliveira, P.G.D., Sousa, J.M.D., Assunção, D.G.F., Araujo, E.K.S.D., Bezerra, D.S., Dametto, J.F.D.S. and Ribeiro, K.D.D.S., 2022. Impacts of consumption of ultra-processed foods on the maternal-child health: a systematic review. Frontiers in nutrition, 9, p.821657.
Piñeiro, S.A. and Cerniglia, C.E., 2021. Antimicrobial drug residues in animal‐derived foods: Potential impact on the human intestinal microbiome. Journal of Veterinary Pharmacology and Therapeutics, 44(2), pp.215-222.
Rivera-Chávez, F., Lopez, C.A. and Bäumler, A.J., 2017. Oxygen as a driver of gut dysbiosis. Free Radical Biology and Medicine, 105, pp.93-101.
Rogers, A.W., Tsolis, R.M. and Bäumler, A.J., 2021. Salmonella versus the Microbiome. Microbiology and Molecular Biology Reviews, 85(1), pp.10-1128.
Sender, R., Fuchs, S. and Milo, R., 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS biology, 14(8), p.e1002533.
Securing the future of microbiome research and innovation: the need for biobanking infrastructure in the UK Report: cabidigitallibrary.org/doi/full/10.5555/20240382445
Shivakoti, R., Biggs, M.L., Djoussé, L., Durda, P.J., Kizer, J.R., Psaty, B., Reiner, A.P., Tracy, R.P., Siscovick, D. and Mukamal, K.J., 2022. Intake and sources of dietary fiber, inflammation, and cardiovascular disease in older US adults. JAMA network open, 5(3), pp.e225012-e225012.
The UK Science and Technology Framework: The UK Science and Technology Framework: taking a systems approach to UK science and technology
Valdes, A.M., Walter, J., Segal, E. and Spector, T.D., 2018. Role of the gut microbiota in nutrition and health. BMJ, 361.
Abbreviations
In this guide
In this guideAbbreviations
|
Adverse Outcome Pathways |
AOP |
|
Acceptable Daily Intake |
ADI |
|
antimicrobial resistance |
AMR |
|
Committee on Toxicity of Chemicals in Food, Consumer Products and the Environment |
COT |
|
Food Standards Agency |
FSA |
|
Faecal Microbiota Transplant |
FMT |
|
guidance for industry |
GFI |
|
Inflammatory bowel disease |
IBD |
|
Joint FAO/WHO Expert Committee on Food Additives |
JECFA |
|
Joint Meeting on Pesticide Residues |
JMPR |
|
microbiological Acceptable Daily Intake |
mADI |
|
no-observed adverse effect concentrations/levels |
NOAEC/Ls |
|
Quantitative PCR |
qPC |
|
ribosomal RNA |
rRNA |
|
Short chain fatty acids |
SCFAs |
|
T helper 17 cells |
TH17 |
|
Ultra processed foods |
UPF |
|
Veterinary International Conference on Harmonization |
VICH |
Organizing Committee
Dr Olivia Osborne
Ms Cath Mulholland
Ms Sophy Orphanos
Acknowledgements
We wish to acknowledge the UK Food Standards Agency (UK FSA), the Committee on Toxicity of Chemicals in Food, Consumer Products and the Environment (COT) and participants of the workshop to thank them for their contributions.
Disclaimer
The content of this article does not necessarily reflect the views of the Food Standards Agency or the Committee on Toxicity of Chemicals in Food, Consumer Products and the Environment, nor should their endorsement of any of the content be inferred.
This article contains summaries of the presentations and discussions that took place in the workshop. As such, they represent the views of the authors, and no official endorsement from the participants, either as an individual or as the company/institution/agency that they represent.
COT Members
Professor Alan Boobis (Chair)
Dr Stella Cochrane
Dr James Coulson
Dr Silvia Gratz
Professor Thorhallur I. Halldorsson
Professor Gary Hutchison
Professor Gunter Kuhnle
Dr David Lovell
Professor Shirley Price
Dr Mac Provan
Dr Michael Routledge
Dr Cheryl Scudamore
Dr Natalie Thatcher
Professor Mireille Toledano
Dr Simon Wilkinson
Professor Philippe Wilson
Professor Peter Barlow
Dr Steve Enoch
Dr Alison Yeates
Dr Meera Cush
Mr Gordon Burton
Dr Andreas Kolb
Mr Nick Richardson
Dr Chris Morris
Professor Maged Younes
COT Secretariat
Ms Catherine Mulholland
Ms Britta Gadeberg
Dr Alexander Cooper
Dr Barbara Doerr
Ms Jocelyn Frimpong Manso
Mr Barry Maycock
Dr Olivia Osborne
Ms Claire Potter
Ms Sabrina Thomas
Ms Chara Tsoulli
Ms Frederique Uy
Ms Sophy Orphanos
Dr Gaetana Spedalieri
Mr Thomas Hornsby
Ms Gail Drummond
Dr Emily Hudson
Ms Natasha Adams
Ms Abigail Smith
Dr Katie Schulz
Ms Katie Wetherall
Dr Rachel Kerr
Ms Polly Bevan
Ms Alba Ureña Rusillo